
Peter S. Lomdahl
Theoretical Division
Los Alamos National Laboratory
Los Alamos, New Mexico 87545
pxl@lanl.gov
(Presented at the 6th International Python Conference, San Jose, California. October 14-17, 1997).
While the development of SPaSM is an ongoing effort, we have been hampered by a number of serious problems. First, typical simulations generate tens to hundreds of gigabytes of data that must be analyzed. This task is not easily performed on a user's workstation, nor is it economically feasible to buy everyone their own personal desktop supercomputer. A second problem is that of interactivity and control. We are constantly making changes to investigate new physical models, different materials, and so forth. This would usually require changes to the underlying C code--a process that was tedious and not very user friendly. We wanted a more flexible mechanism. Finally, there were many difficulties associated with the development and maintenance of our software. While we are only a small group, it was not uncommon for different users to have their own private copies of the software that had been modified in some manner. This, in turn, led to a maintenance nightmare that made it almost impossible to update the software or apply bug-fixes in a consistent manner.
To address these problems, we started investigating the use of scripting languages. In 1995, we wrote a special purpose parallel-scripting language, but replaced it with Python a year later (although the interface generation tool for that scripting language lives on as SWIG). In this paper, we hope to describe some of our experiences with Python and how it has helped us solve practical scientific computing problems. In particular, we describe the organization of our system, module building process, interesting tools that Python has helped us develop, and why we think this approach is particularly well-suited for scientific computing research.
Another factor, not to be overlooked, is the increasing acceptance of Python elsewhere in the computational science community. Efforts at Lawrence Livermore National Laboratory and elsewhere were attractive--in particular, we saw Python as being a potential vehicle for sharing modules and utilizing third-party tools [4,9,10].
The major components of our system take the form of C libraries. When Python is used, the libraries are compiled into shared libraries and dynamically loaded into Python as extension modules. The functionality of each library is exposed as a collection of Python "commands." Because of this, the C and Python programming environments are closely related. In many cases, it is possible to implement the same code in both C or Python. For example :
/* A simple function written in C */
#include "SPaSM.h"
void run(int nsteps, double Dt, int freq) {
int i;
char filename[64];
for (i = 0; i < nsteps; i++) {
integrate_adv_coord(Dt);
boundary_periodic();
redistribute();
force_eam();
integrate_adv_velocity(Dt);
if ((i % freq) == 0) {
sprintf(filename,"Dat%d",i);
output_particles(filename);
}
}
}
Now, in Python :
# A function written in Python
from SPaSM import *
def run(nsteps, Dt, freq):
for i in xrange(0, nsteps):
integrate_adv_coord(Dt)
boundary_periodic()
redistribute()
force_eam()
integrate_adv_velocity(Dt)
if (i % freq) == 0) :
output_particles('Dat'+str(i))
Handling two operational modes introduces a number of problems related to code maintenance and installation. Traditionally, we would recompile the entire system and all of its modules for each configuration (placing the files in an architecture dependent subdirectory). Users would have to decide which system they wanted to use and compile all of their modules by linking against the appropriate libraries and setting the right compile-time options. Changing configurations would typically require a complete recompile.
With dynamic loading and shared libraries however, we have been able to devise a different approach to this problem. Rather than recompiling everything for each configuration, we use an implementation independent layer of system wrappers. These wrappers provide a generic implementation of message passing, parallel I/O, and thread management. All of the core modules are then compiled using these generic wrappers. This makes the modules independent of the underlying operational mode---to use MPI or threads, we simply need to supply a different implementation of the system wrapper libraries. This is easily accomplished by building two different versions of Python that are linked against the appropriate system libraries. To run in a particular mode, we now just run the appropriate version of Python (i.e. 'python' or 'pythonmpi'). The neat part about this approach is that all of the modules work with both operational modes without any recompilation or reconfiguration. If a user is using threads, but wants to switch to MPI, they simply run a different version of Python--no recompilation of modules is necessary.
A full discussion of writing system-wrappers can be found elsewhere. In particular, a discussion of writing parallel I/O wrappers for Python can be found in [5]. An earlier discussion of the technique we have used for writing message passing and I/O wrappers can also be found in [7].
// SWIG interface file
%module SPaSM
%{
#include "SPaSM.h"
%}
void integrate_adv_coord(double Dt);
void boundary_periodic();
void redistribute();
void force_eam();
void integrate_adv_velocity(double Dt);
int output_particles(char *filename);
typedef struct {
double x,y,z;
} Vector;
typedef struct {
int type;
Vector r;
Vector v;
Vector f;
} Particle;
// SWIG interface for vector and particle structures
%include datatypes.h
// Extend data structures with a few useful methods
%addmethods Vector {
char *__str__() {
static char a[1024];
sprintf(a,"[ %0.10f, %0.10f, %0.10f ]", self->x, self->y, self->z);
return a;
}
}
%addmethods Particle {
Particle *__getitem__(int index) {
return self+index;
}
char *__str__() {
// print out a particle
...
}
}
When the Python interface is built, C structures now appear like
Python objects. By providing Python-specific methods (such as
__getitem__) we can even provide array access.
For example, the following Python code would print out
all of the coordinates of stored particles :
# Print out all coordinates to a file
f = open("part.data","w")
p = SPaSM_first_particle() # Get first particle pointer
for i in xrange(0,SPaSM_count_particles()):
f.write("%f, %f, %f\n" % (p[i].r.x, p[i].r.y, p[i].r.z))
f.close()
While only a simple example, having direct access to underlying data has
proven to be quite valuable since we view the internal representation
of data, check values, and perform diagnostics.
One of Python's most powerful features is its exception handling mechanism. Exceptions are easily raised and handled in Python scripts as follows :
# A Python function that throws an exception
def allocate(nbytes):
ptr = SPaSM_malloc(nbytes)
if ptr == "NULL": raise MemoryError,"Out of memory!"
# A Python function that catches an exception
def foo():
try:
allocate(NBYTES)
except:
return # Bailing out
/* A C function that throws an exception */
void *SPaSM_malloc(size_t nbytes) {
void *ptr = (void *) malloc(nbytes);
if (!ptr) Throw("SPaSM_malloc : Out of memory!");
return ptr;
}
/* A C function catching an exception */
int foo() {
void *p;
Try {
p = SPaSM_malloc(NBYTES);
} Except {
printf("Unable to allocate memory. Returning!\n");
return -1;
}
}
In the case of a stand-alone C application, exceptions can
be caught and handled internally. Should an uncaught exception
occur, the code prints an error message and terminates. However,
when Python is used, we can
generate Python exceptions using a SWIG user-defined
exception handler such as the following :
%module SPaSM
// A SWIG user defined exception handler
%except(python) {
Try {
$function
} Except {
PyErr_SetString(PyExc_RuntimeError,SPaSM_error_msg());
}
}
// C declarations
...
The handler code gets placed into all of the Python
"wrapper" functions and is responsible for translating C exceptions
into Python exceptions. Doing this makes our physics code operate
in a more seamless manner and gives it a precisely defined error recovery
procedure (i.e. internal errors always result Python exceptions). For example :
We have found error recovery to be critical. Without it, simulations may continue to run, only to generate wrong answers or a mysterious system crash.>>> SPaSM_malloc(1000000000) RuntimeError: SPaSM_malloc(1000000000). Out of memory! (Line 52 in memory.c) >>>
void plot_spheres(Plot3D *p3, DataFunction func, double min, double max,
double radius) {
Particle *p = Particles;
int i, npart, color;
npart = SPaSM_count_particles();
for (i = 0; i < npart; i++, p++) {
/* Compute color value */
value = (*func)(p);
color = (value-min)/(max-min)*255;
/* Plot it */
Plot3D_sphere(p3,p->r.x,p->r.y,p->r.z,radius,color);
}
}

While simple, we often want our images to contain more information including titles, axis labels, colorbars, time-stamps, and bounding boxes. To do this, we have built an object-oriented visualization framework in Python. Python classes provide methods for graph annotation and common graph operations. If a user wants to make a new kind of plot, they simply inherit from an appropriate base class and provide a function to plot the desired data. For example, a "Sphere" plot using the above C function would look like this :
class Spheres(Image3D):
def __init__(self, func, min, max, radius=0.5):
Image3D.__init__(self, ...
self.func = func
self.min = min
self.max = max
self.radius = radius
def draw(self):
self.newplot()
plot_spheres(self.p3,self.func,self.min,self.max,self.radius)
>>> s = Spheres(PE,-8,-3.5) # Create a new image >>> s.rotd(45) # Rotate it down >>> s.zoom(200) # Zoom in >>> s.show() # generate Image and display it >>> ...
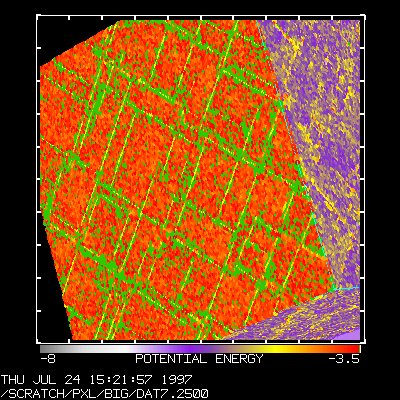
Thus, with a simple C function and a simple Python class, we get a lot of functionality for free, including methods for image manipulation, graph annotation, and display. In the current system, there are about a dozen different types of images for 2D and 3D plotting. It is also possible to perform filtering, apply clipping planes, and other advanced operations. Most of this is supported by about 2000 lines of Python code and a relatively small C library containing performance critical operations.
# Simplified script using a web-server
from vis import *
from web import *
web = SPaSMWeb()
web.add(WebMain(""))
web.add(WebFileText("Msg"+`run_no`))
# Create an image object
set_graphics_mode(HTTP)
ke = Spheres(KE,0,20)
ke.title = "Kinetic Energy"
web.add(WebImage("ke.gif",ke))
def run(nsteps):
for i in xrange(0,nsteps):
integrate(1) # Integrate 1 timestep
web.poll() # Anyone looking for me?
# Run it
run(10000)
While simple, this example gives the basic idea. We create a
server object and register a few "links." When a user connects with
the server, they will be presented with some status information and a
list of available links. When the links are accessed, the server
feeds real-time data back to the user. In the case of
images, they are generated immediately and sent back in GIF format. During
this process, no temporary files are created, nor is the server
transmitting previously stored information (i.e. everything is
created on-the-fly).
While we are still refining the implementation, this approach has turned out to be useful--not only can we periodically check up on a running simulation, this can can be done from any machine that has a Web-browser, including PCs and Macs running over a modem line (allowing a bored physicist to check on long-running jobs from home or while on travel). Of course, the most amazing fact of all is that this was implemented by one person in an afternoon and only involved about 150 lines of Python code (with generous help from the Python library).
We have also used Python in conjunction with code management. One of the newer problems we have faced is the task of finding the definitions of C functions (for example, we might want to know how a particular command has been implemented in C). To support this, we have written tools for browsing source directories, displaying definitions, and spawning editors. All of this is implemented in Python and can be performed directly from the Physics application. Python has made these kinds of tools very easy to write--often only requiring a few hours of effort.
One point, that we would like to strongly emphasize, is the dynamic nature of many scientific applications. Our application is not a huge monolithic package that never changes and which no one is supposed to modify. Rather, the code is constantly changing to explore new problems, try new numerical methods, or to provide better performance. The highly modular nature of Python works great in this environment. We provide a common repository of modules that are shared by all of the users of the system. Different users are typically responsible for different modules and all users may be working on different modules at the same time. Whenever changes to a module are made, they are immediately propagated to everyone else. In the case of bug-fixes, this means that patched versions of code are available immediately. Likewise, new features show up automatically and can be easily tested by everyone. And of course, if something breaks--others tend to notice almost immediately (which we feel is a good thing).
A second problem is that of education--we have found that some users have a very difficult time making sense of the system (even we're somewhat amazed that it really works). Interestingly enough, this does not seem to be directly related to the use of C code or Python for that matter, but to issues related to the configuration, compilation, debugging, and installation of modules. While most users have written programs before, they have never dealt with shared libraries, automated code generators, high-level languages, and third-party packages such as Python. In other cases, there seems to be a "fear of autoconf"--if a program requires a configure script, it must be too complicated to understand (which is not the case here). The combination of these factors has led some users to believe that the system is far more complicated and fragile than it really is. We're not sure how to combat this problem other than to try and educate the users (i.e. it's essentially the same code as before, but with a really cool hack).
[1] D.M.Beazley and P.S. Lomdahl, "Message Passing Multi-Cell Molecular Dynamics on the Connection Machine 5," Parallel Computing 20 (1994), p. 173-195.
[2] P.S.Lomdahl, P.Tamayo, N.Gronbech-Jensen, and D.M.Beazley, "50 Gflops Molecular Dynamics on the CM-5," Proceedings of Supercomputing 93, IEEE Computer Society (1993), p.520-527.
[3] S.J.Zhou, D.M. Beazley, P.S. Lomdahl, B.L. Holian, Physical Review Letters. 78, 479 (1997).
[4] P. Dubois, K. Hinsen, and J. Hugunin, Computers in Physics (10), (1996) p. 262-267.
[5] D. M. Beazley and P.S. Lomdahl, "Extensible Message Passing Application Development and Debugging with Python", Proceedings of IPPS'97, Geneva Switzerland, IEEE Computer Society (1997).
[6] T.-Y.B. Yang, D.M. Beazley, P. F. Dubois, G. Furnish, "Steering Object-oriented Computations with Python", Python Workshop 5, Washington D.C., Nov. 4-5, 1996.
[7] D.M. Beazley and P.S. Lomdahl, "High Performance Molecular Dynamics Modeling with SPaSM : Performance and Portability Issues," Proceedings of the Workshop on Debugging and Tuning for Parallel Computer Systems, Chatham, MA, 1994 (IEEE Computer Society Press, Los Alamitos, CA, 1996), pp. 337-351.
[8] D.M. Beazley, "Using SWIG to Control, Prototype, and Debug C Programs with Python", Proceedings of the 4th International Python Conference, Lawrence Livermore National Laboratory, June 3-6, 1996.
[9] K. Hinsen, The Molecular Modeling Toolkit, http://starship.skyport.net/crew/hinsen/mmtk.html
[10] Jim Hugunin, Numeric Python, http://www.sls.lcs.mit.edu/jjh/numpy
[11] Fredrik Lundh, Python Imaging Library, http://www.python.org/sigs/image-sig/Imaging.html